
Ph.D. Computer Science (Texas State University - Anticipated 2024)
M.S. Computer Science (Texas State University - 2020)
M.S. Biochemistry (Texas State University - 2014)
B.S. Biology with minor in Chemistry (Texas State University - 2012)

I previously attended Texas State University from 2008 to 2014, earning both a Bachelor’s of Science in Biology with a minor in Chemistry and a Masters of Science in Biochemistry. Shortly after graduating, I began working at a well-established company known as Grifols, a company that specializes in blood banking and plasma-derived pharmaceuticals. This was my first introduction into what the science industry was like in the modern world. Never-before had I been exposed to such an environment; my mother, who is now retired, worked as a guidance counselor for a school district and my father, now a retired mathematics teacher at a San Antonio High School formerly worked with manufacturing plants dealing with audio and electrical equipment. Furthermore, my sister worked towards becoming an elementary school music teacher and my brother-in-law is a high school band director. It is easy to see that I have always been surrounded by academia, and most of my influential figures also tried motivating me to move towards a career in education or academia.

Even though I had begun my life in the work-force, I knew my journey had yet to reach its conclusion--I have returned to school because I want to combine what I know about science and technology. To illustrate how a grander vision has already begun, I can actually say that I own a business known as “AminoAcidStudios”, currently only hosting a website where students can learn their amino acids via an interactive program. Though this is a small and simple manifestation, it sets the foundation to what will eventually be a greater application of my capabilities, and this has the potential to be a multitude of possibilities. For example, I could design the next platform that is the basis for a particular diagnostic assay, or I could continue on the idea that is AminoAcidStudios and expand on my website to be a resource for the future generations of scientists. This educational undertaking is yet another step in my journey that leads to the manifestation of a grander vision.
I am also a musician (Alto Saxophone and Piano, but I currently mostly dabble with electronic music), I spend my time coding my own side projects, and from time to time, enjoy the occasional chess match.
"PSAAP3" - Department of Energy's Predictive Science Academic Alliance Program
Oden Institute's "PECOS" - The Center for Predictive Engineering and Computational Science
The PARLA orchestration language
Oden Institute - The Center for Predictive Engineering and Computational Science
"The Center for Predictive Engineering and Computational Science is a research center in the Oden Institute for Computational Engineering and Sciences. It is dedicated to the development and application of tools and techniques for making reliable computational predictions of complex systems."
The PECOS Center aims at providing high-fidelity simulations of a plasma torch. I am currently working on parallelizing radiation transport “sweeps” computations for GPUs, which represent an important component of the plasma torch simulation. This parallel implementation will be studied using Parla, an orchestration language for running numerical simulations on heterogenous architectures--Parla automates data movement and kernel calls across all the CPUs and GPUs available on a given machine.
Efficient Computing Laboratory (ECL)
Texas State University - Efficient Computing Laboratory
I am also working on a new GPU parallelization method for computing Minimum Spanning Trees (MSTs).
A Parallel Implementation of a Greedy TSP Algorithm
The “Traveling Salesman Problem” (TSP) is a puzzle-like rendition of a question that simply poses “Which is the shortest route that a salesperson can take to visit all cities on a map exactly once and return to the starting city while keeping the distance of the route at a minimum?” It is a Combinatorial Optimization Problem that is often used as an exploratory topic for finding better solutions to other COPs. Formally, the way of describing the route the salesman takes is defined as a “Hamiltonian Path,” where every vertex of a complete, unweighted graph is visited exactly once before returning to the starting vertex.
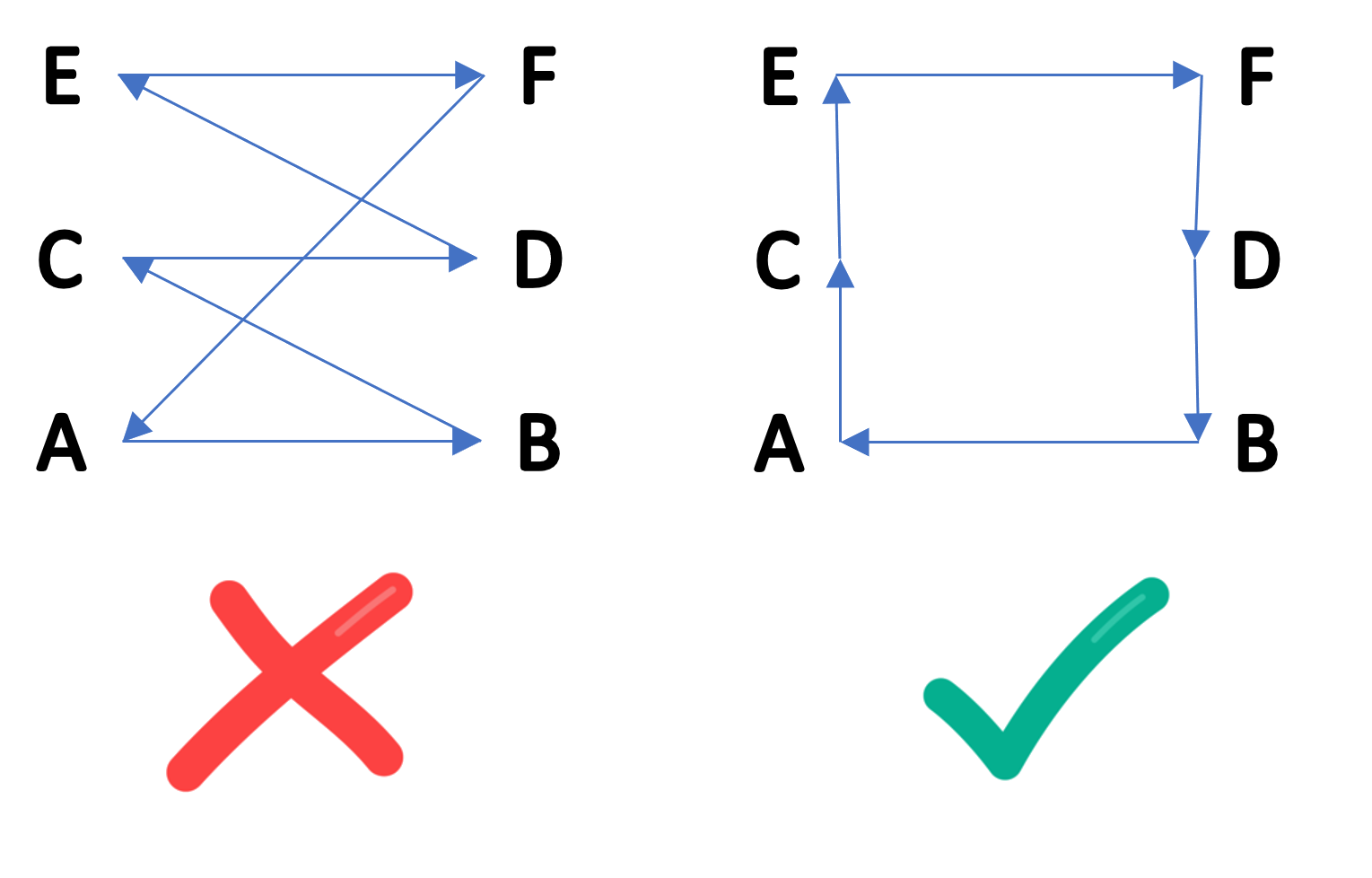
Finding the optimal TSP solution is NP-hard and, therefore, intractable for large problem sizes. This is why many heuristics approaches have been proposed for finding near-optimal solutions in polynomial time. One particularly good heuristic is a greedy approach based on Kruskal's algorithm, a minimum-spanning tree algorithm that uses sorted edge weights to connect vertices. It attempts to generate a minimal tour by using the edges with the least weights while following two simple rules: (1) a vertex cannot have a degree of 3 or more and (2) adding an edge to the tree cannot create a cycle unless the edge to be added is the last edge connecting the initial and final vertices.
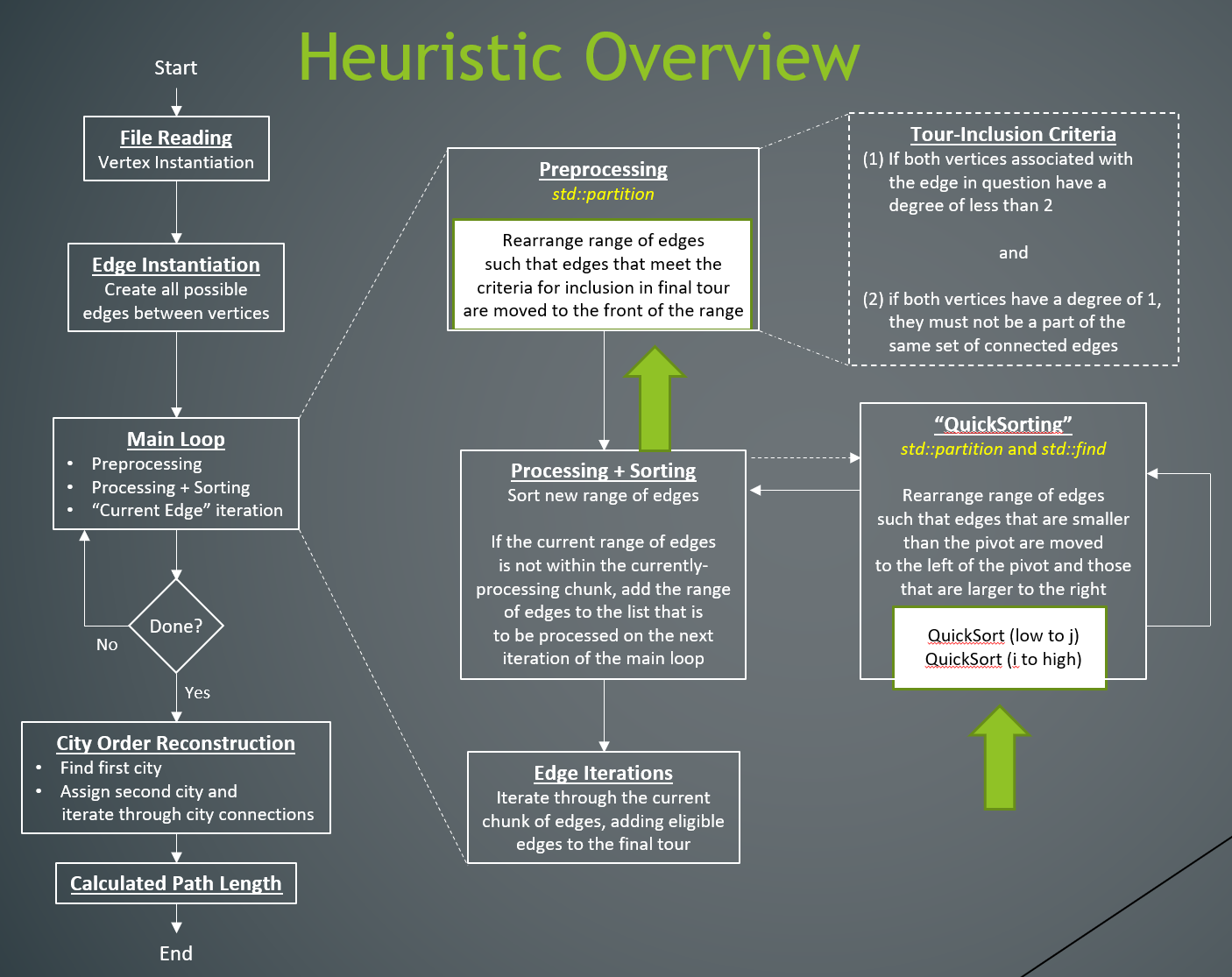
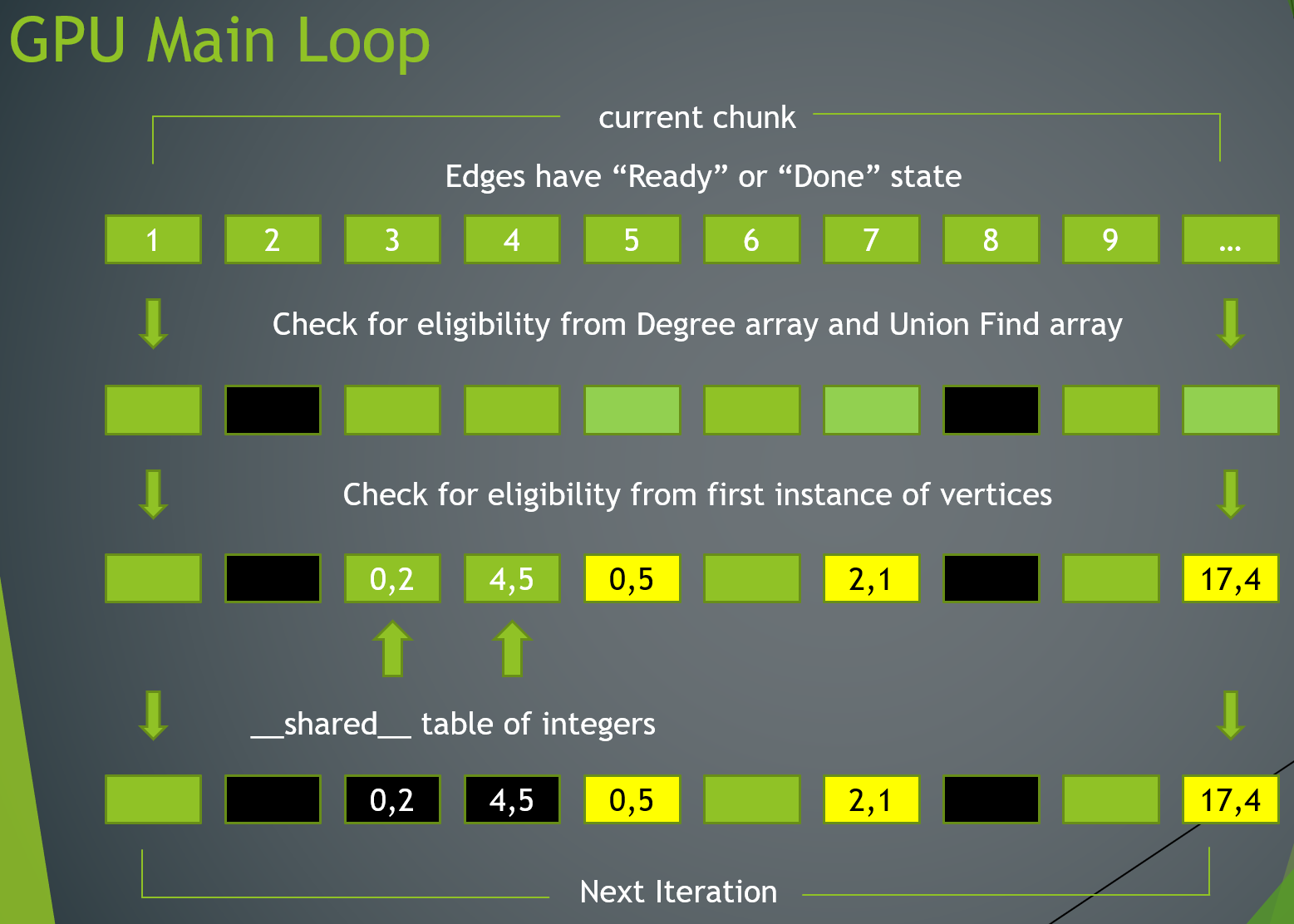
My thesis research focused on parallelizing this greedy TSP algorithm to boost its performance. The first step—sorting the edges—is relatively easy to accomplish in parallel. However, the second step—adding edges according to the two rules—had never before been successfully parallelized due to the intricate nature of the dependencies. Whether an edge must be included depends entirely on which lighter-weighted edges have already been included, meaning that a direct parallelization of the algorithm is not possible. However, after careful evaluation of the algorithm, the implementation made light of the simple fact that all vertices eventually need to be included in the final path, meaning edges could be considered independently of each other (after sorting) so long as no edges that had those vertices had yet to be included in the final path. This new approach aimed to extract as much parallelism as possible, was implemented in the CUDA language for GPU execution, and achieved speedups that were significantly more efficient than other algorithms.
Binding Affinity of Epithelial Sodium Channel Subunits and Peptides Measured Through Surface Plasmon Resonance (2014)
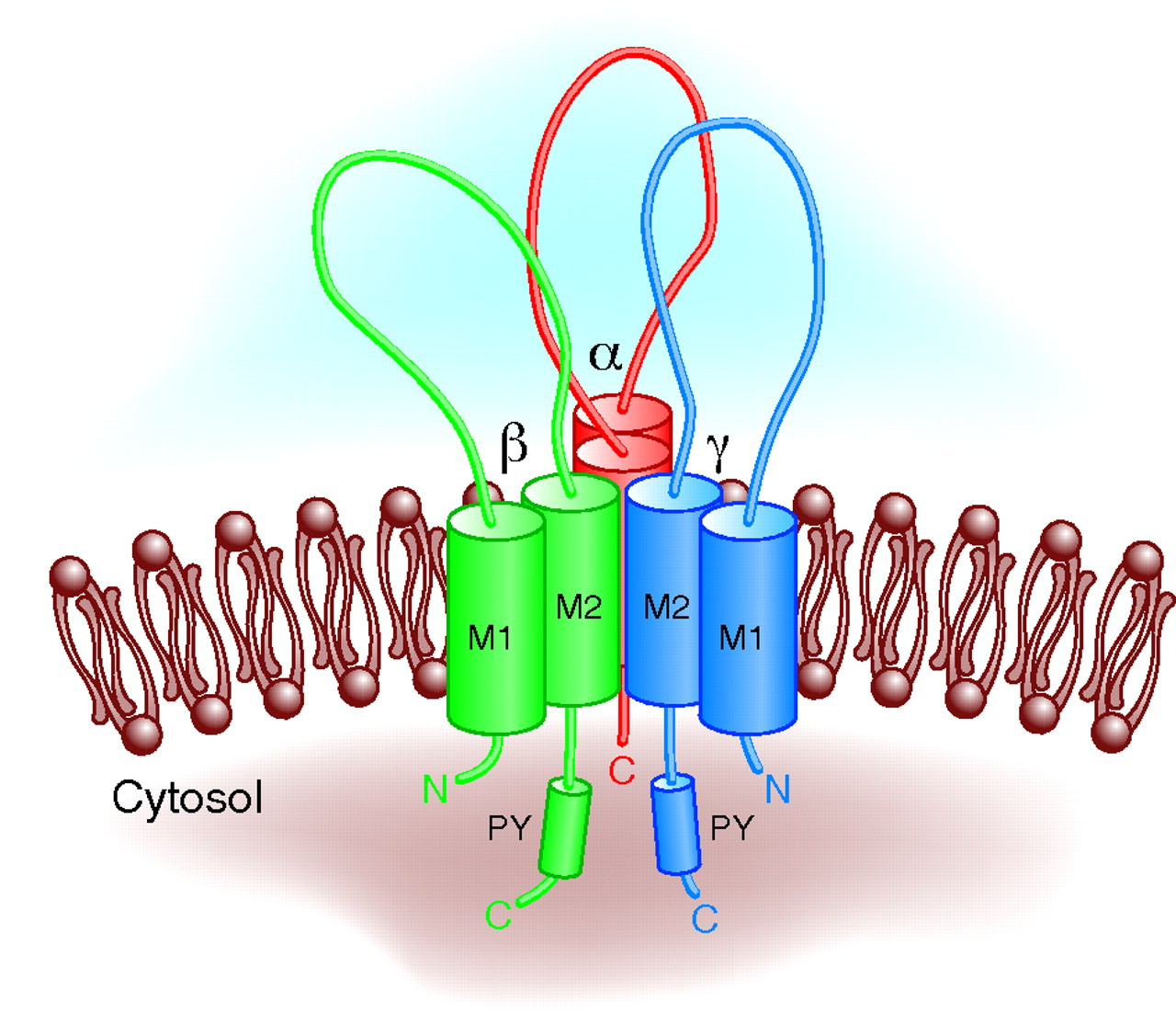
Salt levels play a crucial role in the regulation of blood pressure, and cells are able to maintain salt levels via ion channels embedded within the plasma membrane. The Epithelial Sodium Channel (ENaC) is a heterotrimeric membrane protein that allows the passage of sodium, and this study’s aim is to further understand what structural aspects of ENaC are responsible for maintaining proper function of the channel. Potential residues critical for interaction between ENaC alpha, beta, and gamma subunits were identified, and peptide segments containing the targeted residues were PCR amplified, ligated onto a pGEX-4t-2 vector, and transformed into Top10 expression E. Coli. This vector contains a GST-tag that was used to purify the peptides and immobilize them for further analysis by surface plasmon resonance (SPR).
.png)
Binding between sets of peptides predicted to be important for stabilizing interactions in ENaC was investigated using SPR. One peptide from a potential interacting set of peptide regions was immobilized through the GST-tag, and binding with the corresponding partner (with the GST-tag removed) was quantified. Two interactions—one between the alpha-palm and beta-thumb, and the other between the beta-palm and gamma-thumb regions of ENaC—were analyzed. The alpha-palm and beta-thumb interaction had a measured Kd of 2.9 x 10-8 M, whereas the interaction from the beta-palm and gamma-thumb regions was in the micromolar range. Additional sets of peptides have been expressed and will be analyzed for binding interactions. Results obtained from this initial study will be compared to those obtained from a mutation screen of ENaC function to confirm whether the predicted residues play a crucial structural role related to function.
Gonzalez, A. and Burtscher, M. (2020) A PARALLEL IMPLEMENTATION OF A GREEDY TSP ALGORITHM [Masters Thesis, Texas State University] Efficient Computing Laboratory
Gonzalez, A. and Booth, R. (2014) BINDING AFFINITY OF EPITHELIAL SODIUM CHANNEL SUBUNITS AND PEPTIDES MEASURED THROUGH SURFACE PLASMON RESONANCE [Masters Thesis, Texas State University] Department of Chemistry and Biochemistry
Fall 2021 - CS1319 Foundations of Computer Science
Texas State Email: ag1548@txstate.edu
Efficient Computing Laboratory (ECL)
Texas State University - Department of Computer Science
Texas State University - Department of Chemistry and Biochemistry
PSAAP3 - Predictive Science Academic Alliance Program
Oden Institute's "PECOS" - The Center for Predictive Engineering and Computational Science